Genetics 1D
DNA Replication
Every living organism must make a copy of its chromosomes and hence genes in each cell every time a cell divides. This fidelity of replication must be both rapid and accurate in order to guarantee the organism’s survival. The very structure of DNA ensures that this replication process is accurate. In the next three pages DNA Replication is illustrated. However, because of its complexity any description can only be considered to be highly simplified!
Every living organism must make a copy of its chromosomes and hence genes in each cell every time a cell divides. This fidelity of replication must be both rapid and accurate in order to guarantee the organism’s survival. The very structure of DNA ensures that this replication process is accurate. In the next three pages DNA Replication is illustrated. However, because of its complexity any description can only be considered to be highly simplified!
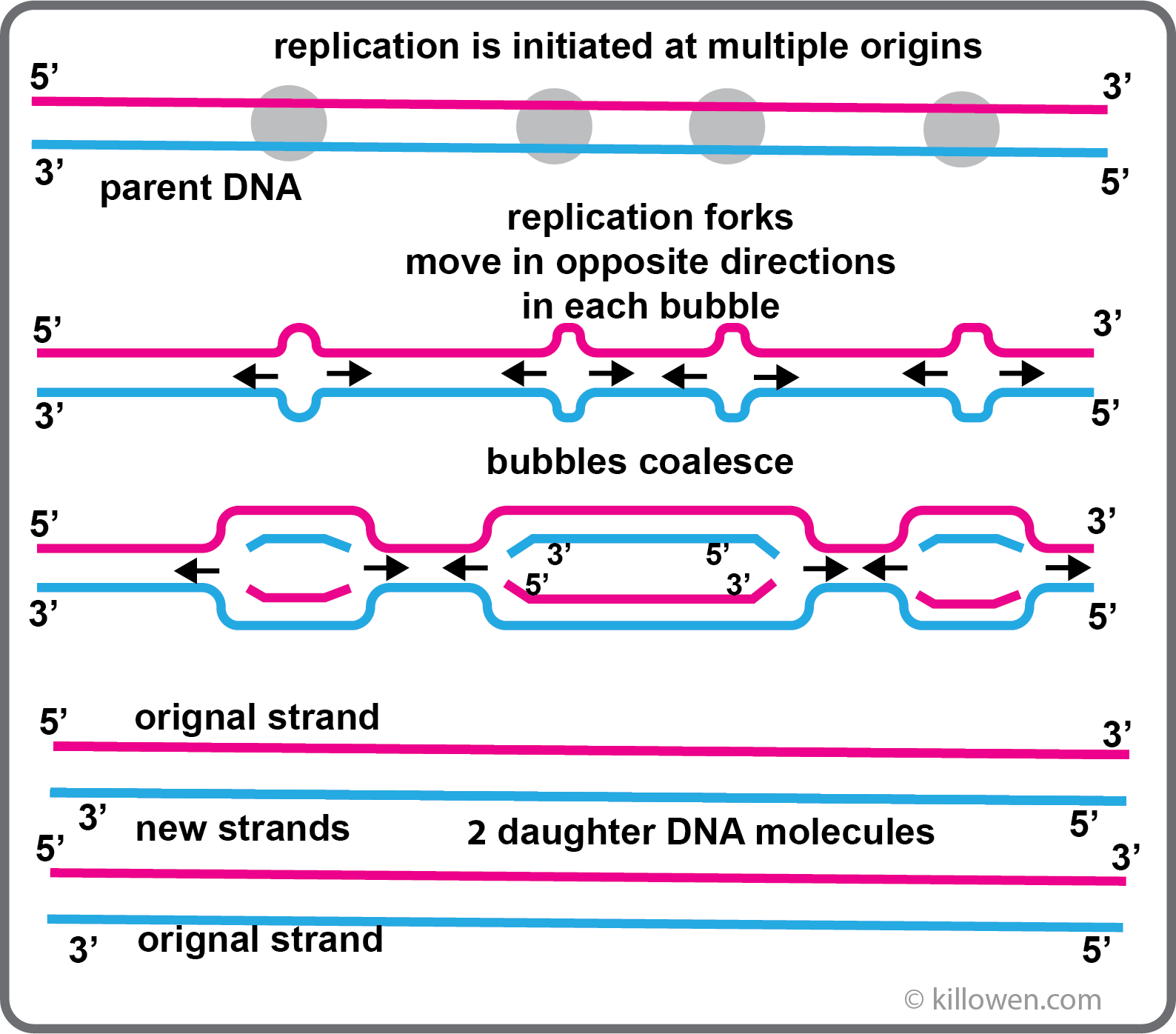
DNA Replication takes place when a cell divides producing two identical daughter cells each containing the same number of chromosomes as the original cell - mitosis.
Each strand of DNA in the parent cell acts as a template for the synthesis of a new complementary strand. This replication process is semi-conservative; the daughter cell contains one strand from the original molecule and a newly formed strand.
If replication just started at one end of the chromosome and continued in a continuous manner to the end, the process would take far too long (30 days is a rough estimation). Instead DNA replication is initiated at multiple sites along the chromosome by specific enzymes. The multiple origins are about 40 kb apart. As the DNA unzips two replication forks are formed which move in opposite directions within the replication bubble. When all the bubbles have coalesced the result is two daughter DNA molecules; this process takes a matter of minutes.
Each strand of DNA in the parent cell acts as a template for the synthesis of a new complementary strand. This replication process is semi-conservative; the daughter cell contains one strand from the original molecule and a newly formed strand.
If replication just started at one end of the chromosome and continued in a continuous manner to the end, the process would take far too long (30 days is a rough estimation). Instead DNA replication is initiated at multiple sites along the chromosome by specific enzymes. The multiple origins are about 40 kb apart. As the DNA unzips two replication forks are formed which move in opposite directions within the replication bubble. When all the bubbles have coalesced the result is two daughter DNA molecules; this process takes a matter of minutes.
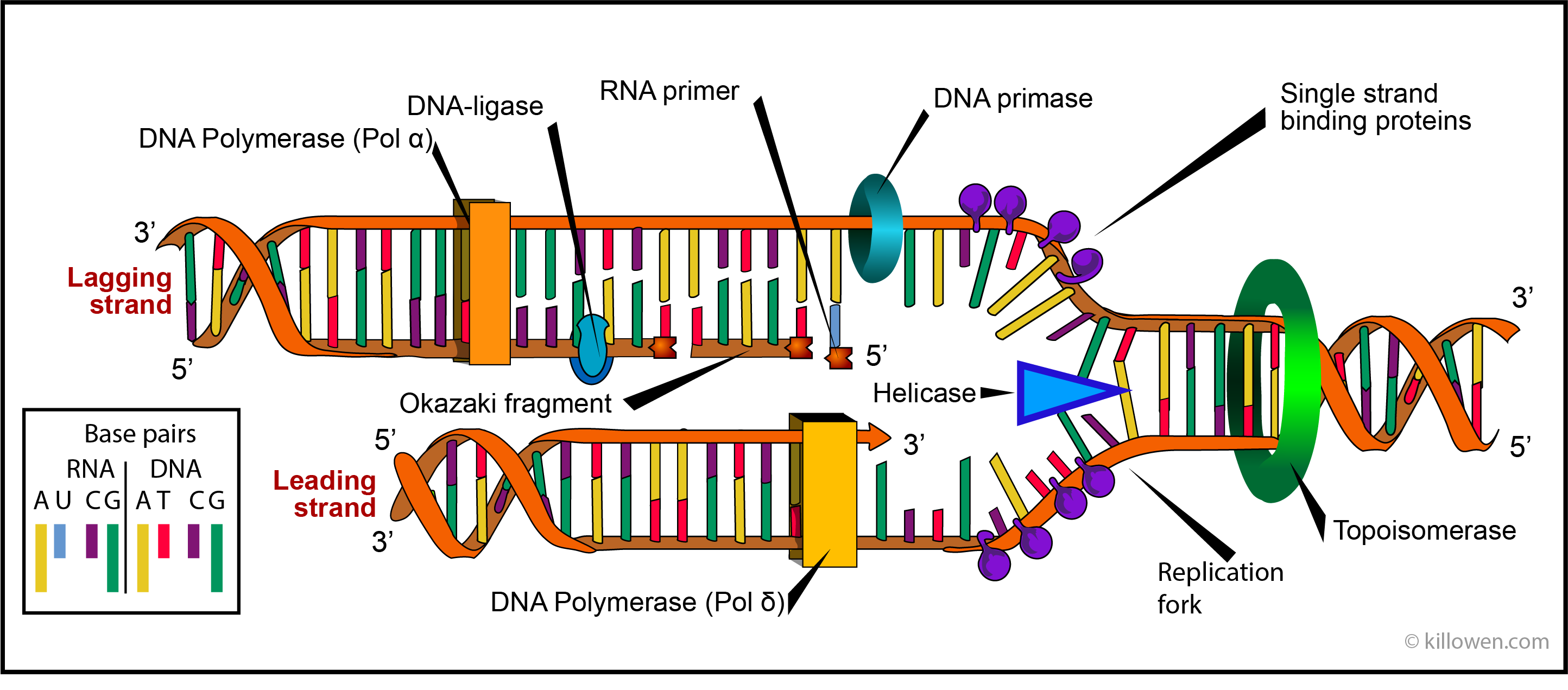
DNA polymerases are enzymes which synthesise or repair DNA. Eukaryotic cells contain 5 DNA polymerases called α, β, γ, δ, and ε. Polymerases α, β, γ, δ, are found in the cell nucleus whereas polymerase ε effects the replication of mitochondria. Polymerase β is active in repairing DNA; polymerases α, β, δ, are active in DNA replication, Polymerases α and δ are shown in the diagram below acting on the lagging and leading stands respectively. It is these polymerases which ensure the accuracy of the DNA replication by choosing the correct nucleotide for inclusion in the ever growing DNA molecule.
Looking at DNA Replication in more detail
The diagram below represents the replication complex which moves along each replication fork. It is important to realise that replication can only take place in the 5’ to 3’ direction on the strand being formed. Since the two strands which form the DNA double helix run in opposite directions, continuous synthesis of a new strand can only take place on one of these strands i.e. on the template which runs in the 3’ to 5’ direction. This is the lower of the two strand shown in the diagram; it is called the leading strand.
The other strand is called the lagging strand. This poses the question how does the new strand form when the template strand runs in the 5’ to 3’ direction. The new strand would have to be synthesised in the opposite direction i.e. 3’to 5’. But this cannot happen. The solution is found when it is realised that this strand is formed in the 5’ to 3’ direction in small discontinuous pieces called Okasaki fragments which are later joined up.
Just ahead of the replication fork the enzyme helicase unwinds the two strands of the parent DNA by breaking the hydrogen bonds holding the base pairs together. Once separated the two single strand regions are stabilized by single-strand binding proteins. These prevent the bases from reannealing before they can be acted on by the polymerase.
As the helicase moves forward the double helix is subjected to a twisting strain. This occurs since DNA replication requires that the double helix is not only unzipped but unwound as well. This unwinding uses the enzymes topoisomerases. These act by forming short breaks in the DNA strands and then rejoining the strands so preventing any strain.
The diagram below represents the replication complex which moves along each replication fork. It is important to realise that replication can only take place in the 5’ to 3’ direction on the strand being formed. Since the two strands which form the DNA double helix run in opposite directions, continuous synthesis of a new strand can only take place on one of these strands i.e. on the template which runs in the 3’ to 5’ direction. This is the lower of the two strand shown in the diagram; it is called the leading strand.
The other strand is called the lagging strand. This poses the question how does the new strand form when the template strand runs in the 5’ to 3’ direction. The new strand would have to be synthesised in the opposite direction i.e. 3’to 5’. But this cannot happen. The solution is found when it is realised that this strand is formed in the 5’ to 3’ direction in small discontinuous pieces called Okasaki fragments which are later joined up.
Just ahead of the replication fork the enzyme helicase unwinds the two strands of the parent DNA by breaking the hydrogen bonds holding the base pairs together. Once separated the two single strand regions are stabilized by single-strand binding proteins. These prevent the bases from reannealing before they can be acted on by the polymerase.
As the helicase moves forward the double helix is subjected to a twisting strain. This occurs since DNA replication requires that the double helix is not only unzipped but unwound as well. This unwinding uses the enzymes topoisomerases. These act by forming short breaks in the DNA strands and then rejoining the strands so preventing any strain.
 Adapted from an image by LadyofHats
Adapted from an image by LadyofHats
A DNA polymerase at the start of the DNA synthesis needs a primer to start the process. On the leading strand only one primer is necessary since here the synthesis is continuous. On the lagging strand the synthesis of DNA is discontinuous and proceeds by the formation of Okazaki fragments. Each fragment requires that an RNA primer is first formed; the Okazaki fragment is then synthesised by DNA polymerase α. A consequence of this action is that there is a join between the Okazaki fragment and the RNA primer. In eukaryotic cell division exonucleases remove the RNA primers and replaced them with DNA. The gap between Okazaki fragments are filled by the action of polymerase δ. DNA ligase then catalyses the formation of a phosphodiester bond between neighbouring Okazaki fragments so forming a continuous new strand of DNA.
It should be noted that uracil takes the place of thymine on the RNA primer.
In addition polymerase δ is associated with two further proteins that load the polymerase firstly onto the primer and then maintains its position with the template. These are called the clamp-loading proteins and sliding-clamp proteins. (For simplicity these are not shown in the diagram).
Telomers are found at the 5’-end of the chromosome. As has already been mentioned these are special repeat sequences of nucleotides whose function is to protect the chromosome during cell division and DNA replication. For example the repeat (TTAGG)n is found in honey bees.
Replication starts at the 5’-end with an RNA primer. These RNA primers when formed along the chromosome are changed to DNA by a DNA polymerase as describe above. However, the polymerase cannot act on the RNA primer when this is at the start of the chromosome. If another mechanism did not occur then the chromosome would gradually decrease in length after each cell division during meiosis. This shortening of the chromosome would then affect gene function over time. The problem is solved by a special a enzyme telomerase. This acts by catalysing the replacement of the RNA by DNA in the absence of a DNA template.
It should be noted that uracil takes the place of thymine on the RNA primer.
In addition polymerase δ is associated with two further proteins that load the polymerase firstly onto the primer and then maintains its position with the template. These are called the clamp-loading proteins and sliding-clamp proteins. (For simplicity these are not shown in the diagram).
Telomers are found at the 5’-end of the chromosome. As has already been mentioned these are special repeat sequences of nucleotides whose function is to protect the chromosome during cell division and DNA replication. For example the repeat (TTAGG)n is found in honey bees.
Replication starts at the 5’-end with an RNA primer. These RNA primers when formed along the chromosome are changed to DNA by a DNA polymerase as describe above. However, the polymerase cannot act on the RNA primer when this is at the start of the chromosome. If another mechanism did not occur then the chromosome would gradually decrease in length after each cell division during meiosis. This shortening of the chromosome would then affect gene function over time. The problem is solved by a special a enzyme telomerase. This acts by catalysing the replacement of the RNA by DNA in the absence of a DNA template.